
Otto Warburg, German biochemist after whom the "Warburg effect" is named.
 Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation - Full paper
Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation - Full paper Reviews on PubMed
Reviews on PubMed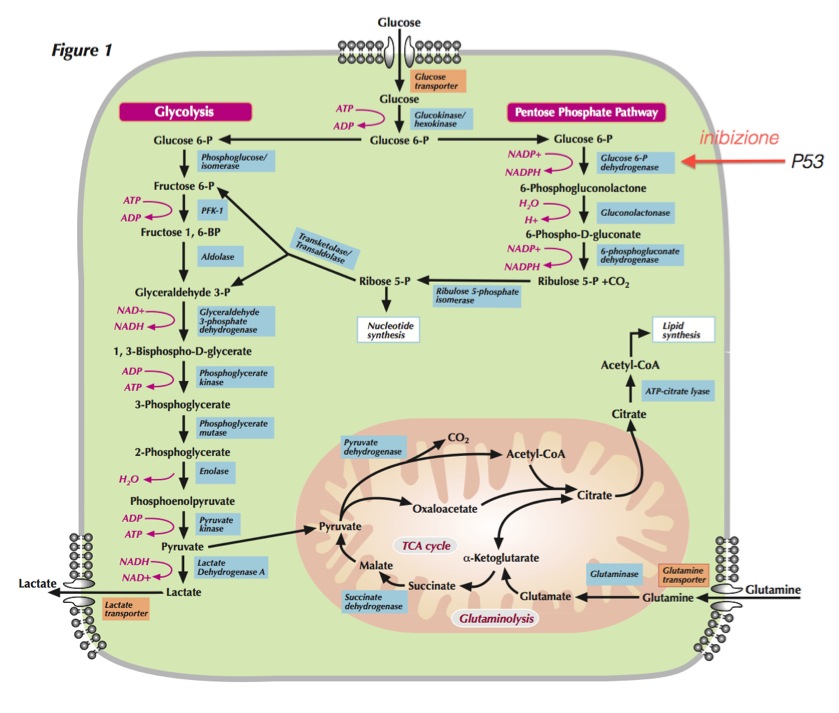 Glycolysis, TCA and PPP metabolism
Glycolysis, TCA and PPP metabolism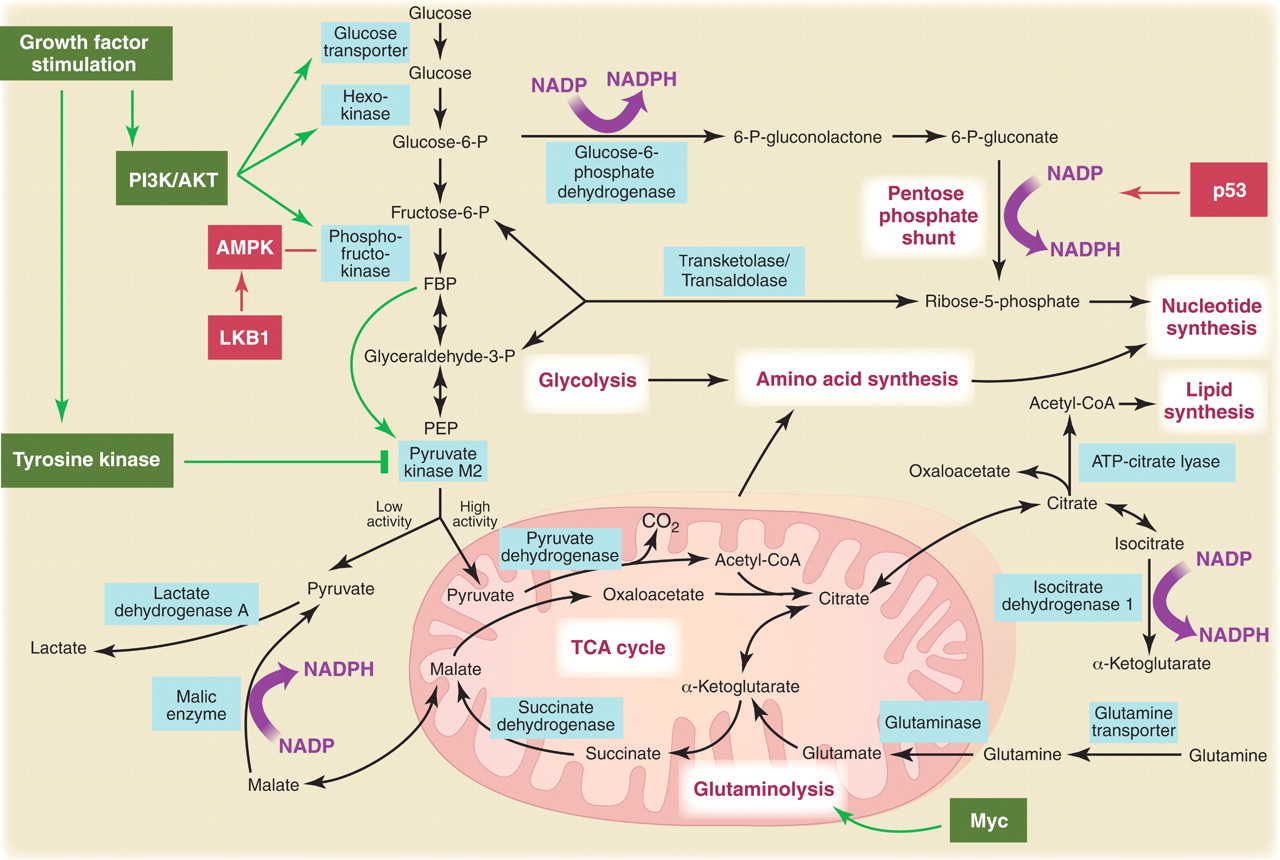
Metabolic pathways active in proliferating cells are directly controlled by signaling pathways involving known oncogenes and tumor suppressor genes. This schematic shows our current understanding of how glycolysis, oxidative phosphorylation, the pentose phosphate pathway, and glutamine metabolism are interconnected in proliferating cells. This metabolic wiring allows for both NADPH production and acetyl-CoA flux to the cytosol for lipid synthesis. Key steps in these metabolic pathways can be influenced by signaling pathways known to be important for cell proliferation. Activation of growth factor receptors leads to both tyrosine kinase signaling and PI3K activation. Via AKT, PI3K activation stimulates glucose uptake and flux through the early part of glycolysis. Tyrosine kinase signaling negatively regulates flux through the late steps of glycolysis, making glycolytic intermediates available for macromolecular synthesis as well as supporting NADPH production. Myc drives glutamine metabolism, which also supports NADPH production. LKB1/AMPK signaling and p53 decrease metabolic flux through glycolysis in response to cell stress. Decreased glycolytic flux in response to LKB/AMPK or p53 may be an adaptive response to shut off proliferative metabolism during periods of low energy availability or oxidative stress. Tumor suppressors are shown in red, and oncogenes are in green. Key metabolic pathways are labeled in purple with white boxes, and the enzymes controlling critical steps in these pathways are shown in blue. Some of these enzymes are candidates as novel therapeutic targets in cancer. Malic enzyme refers to NADP+-specific malate dehydrogenase [systematic name (S)-malate:NADP+ oxidoreductase (oxaloacetate-decarboxylating)].




